- wenqiang.chen@joslin.harvard.edu
- wenqiang.chen.01@regionh.dk
To-Share Protocols
Here I am updating some of my protocols for experiments and analysis. It is a on-going update.

(Co-founder of the Center for Open Science)
- Driven by the concept of open science, I am committed to sharing detailed protocols and methodologies to foster collaboration and accelerate scientific discovery.
- By openly sharing protocols, we can break down barriers to acquire knowledge, enabling researchers worldwide to reproduce and build upon each other’s work with greater ease and accuracy.
- This transparency not only enhances the reliability of scientific research but also democratizes access to new techniques, empowering scientists at all levels to contribute to the advancement of our collective understanding.
- Through this shared approach, we can drive innovation and make meaningful progress in addressing scientific challenges.
Note: some of the protocols are under review or to be published, but I will make that status clear.
(1) Microglia structural analysis
*NOTE: read this JOVE paper I followed to perform this analysis:
Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ
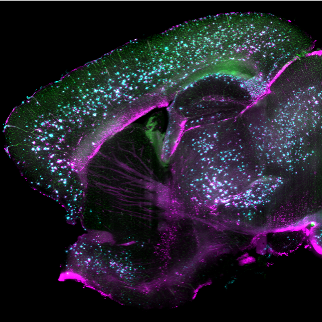
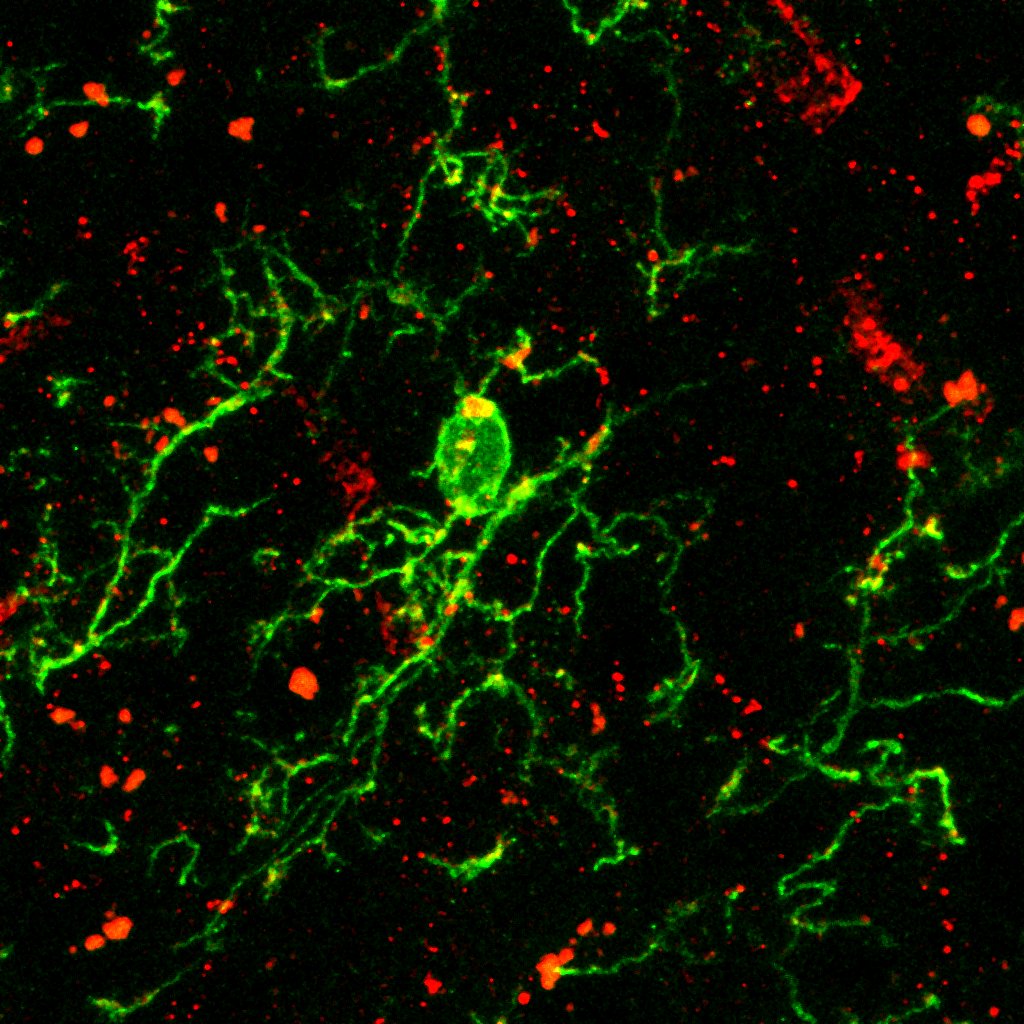
During my teaching ahd demo at the Bordeaux School of Neuroscience (BSN), I showed my students on a relatively advanced analysis on microglial morphological complexity. This is quite useful to microglia and astrocytes, both of which have highly complex structures, and this can play a role in disease pathogenesis (e.g., glial activation).
Below example demonstrated a comparison of microgial structure in different mouse ages, i.e., 3 versus 6 weeks (this is because these are the easiest ages I can request from the organizers at the BSN).
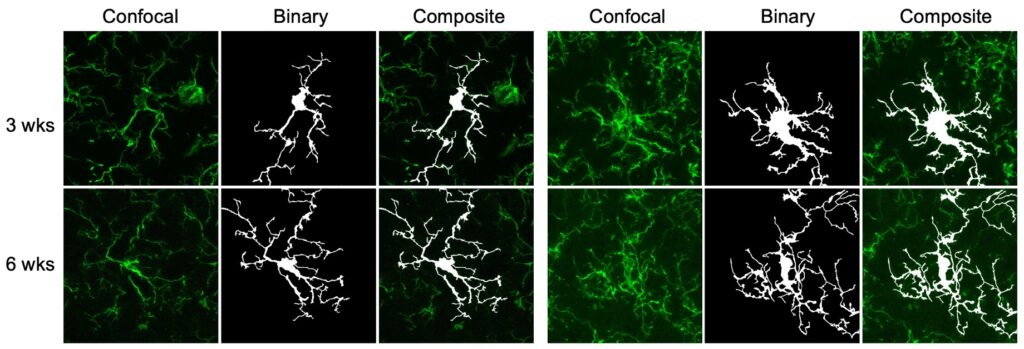
I summarized a detailed protocl to perform this analysis. See photos below.
Tips:
- The most important tip is, you need to acquire good confocal images.
- For Z-stack, do not perform all the stacks. First, you need to make sure you do not stack several cells in the same images.
Further Reading:
Here I would suggest this paper for further reading:
- Paper: https://www.jove.com/t/57648/quantifying-microglia-morphology-from-photomicrographs
- Title: Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ
Descriptions of the papers (to be added here).
(2) Sequencial ultracentrifuge
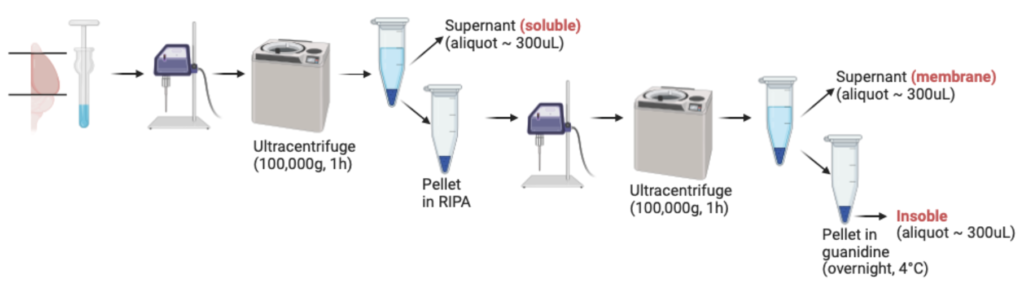
Reference: Loss of insulin signaling in astrocytes exacerbates Alzheimer-like phenotypes in a 5xFAD mouse model
This is one of the most important protocols to extra Abeta plaques from mouse brain hemespheres. I have performed this type of experiment for TOO many times. Very time consuming but very important. Usually, we will save half brain for biochemical analysis, and another half for IHC analysis. Therefore, the mouse can only be perfused with PBS, not further with 4% PFA. See below method I wrote for my manuscript (Chen et al., PNAS. 2023).
For lysates preparation, brain hemispheres were homogenized on ice in 5 volumes (w/v) of 0.1% Triton X-100 in TBS buffer, supplemented with protease inhibitor cocktail (EDTA free, 100X in DMSO, B14002, Bimake, TX, USA) and phosphatase inhibitor cocktail (B15002, 100X, Bimake, TX, USA). After sonication, homogenates were centrifuged at 100,000g for 1 hr at 4.C. The resulting supernatants represented the soluble enriched fraction. The pellets were re-suspended in 1 mL 1x RIPA extraction buffer (20-188, Millipore, MO, USA) supplemented with protease and phosphatase inhibitor cocktails. After sonication, homogenates were centrifuged at 100,000g for 1 hr at 4.C. The resulting supernatants represented the membrane enriched and detergent-soluble fractions. The pellets were resuspended in 1 mL 5 M guanidine-HCl (AAJ6078622, Fisher Scientific, MA, USA) solution overnight at 4.C and centrifuged at 20,000g for 1 hr at 4.C. The resulting supernatants represented the insoluble fraction. Protein concentrations were determined using Pierce BCA Protein Assay Kit (23227, ThermoFisher, MA, USA) prior to biochemical analysis.
Tips:
- blood should be removed by perfusion completely, but this should be fast.
- Homogenization: using douncer, but it has to be pre-cold. I always fixed how many strokes to make this procedure very consistent.
- Ultracentrifugation is very scary. Be care of the balancing.
- Sonication, should be based on piloting experiment. I always performed this in cold room. Very important to keep the tissue on ice or in cold room. Remember to wear enough clothes.
- Resuspension with guanidine-HCl: the mix will get very blurry. Even after centrifugation, be careful there is a dirty layer, try to minimize touching it.
(3) Percoll isolation of microglia
Further reading:
- https://www.jove.com/v/202310/brain-isolation-from-neonatal-mice-for-primary-microglia-culture
- https://www.jove.com/v/63511/isolation-mouse-primary-microglia-magnetic-activated-cell-sorting
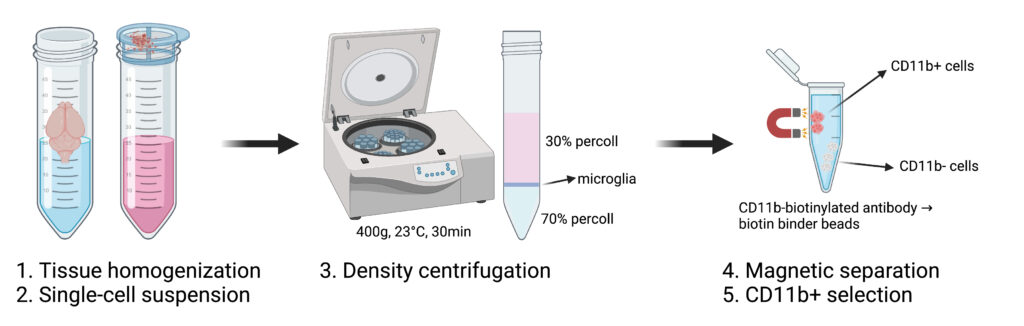
This protocol is used to obtain highly purified microglia from adult mouse brains. So it is primary culture and should be handeled in cell culture hood for primary culture (not to comtaminate your cell lines).
See the protocol I wrote for this purpose, which is also unpublished.
Isolation of primary mouse microglia was previously described (Boroujerdi et al., 2014; Milner et al., 2022; Tamashiro et al., 2012). Briefly, we isolated primary microglia cells from 8-week male IRf/f mouse. Following removal of the brain and the meninges attached on the surface, brain cortices were minced in iced Hybernate A medium (A1247501, ThermoFisher). Tissues were filtered by gentle pipetting and incubated with a digestion buffer that contain a mixture of papain and DNase at 37°C for 30 min. The cell suspension was filtered through a 70 μm filter to remove myelin and cell debris. We used a 30/70 Percoll gradient centrifugation to achieve complete myelin removal. The intermediate layer of the two phases was retrieved and purified with Cd11b+ beads (Miltenyi Biotech). Cd11b+ cells were resuspended in microglia medium (#1901, ScienCell) in 24-well plates. The isolated primary microglia were maintained at 37°C with 5% CO2/95% O2. Medium was changed every other day until reaching confluence for downstream assays.
Tips:
- remove the meninges on the mouse brain.
- probably I would highlight, as always, that percoll centifugation at a low speed, and low acceleration and deceleration (brakes), usually set it as 1, for both.
- Percoll works best at 23 ºC.
- using a long-niddle syringe (3-mL) to underlay 70% percoll
- collecting the 30%/70% phase is very important. Very steady hands are needed.
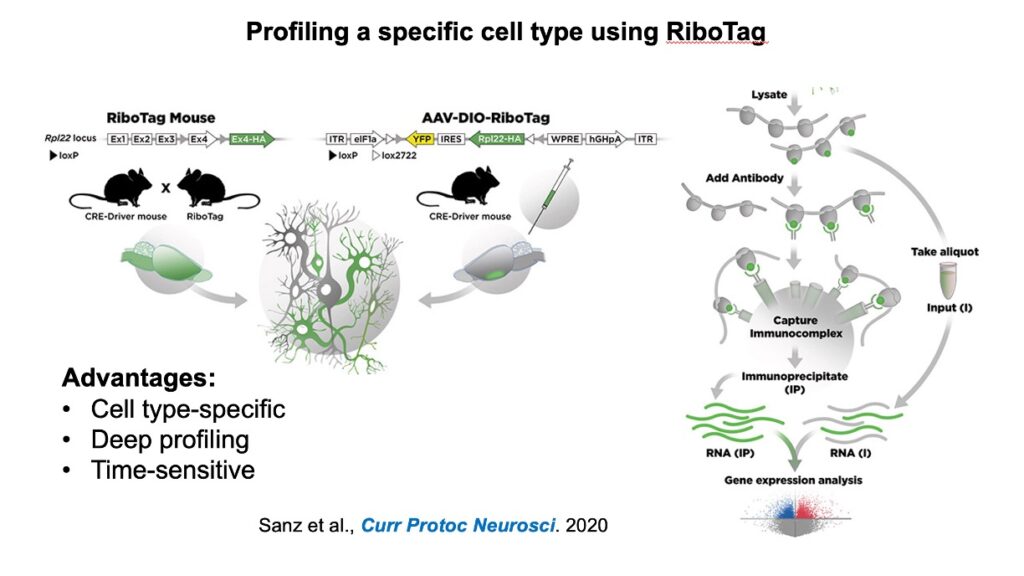
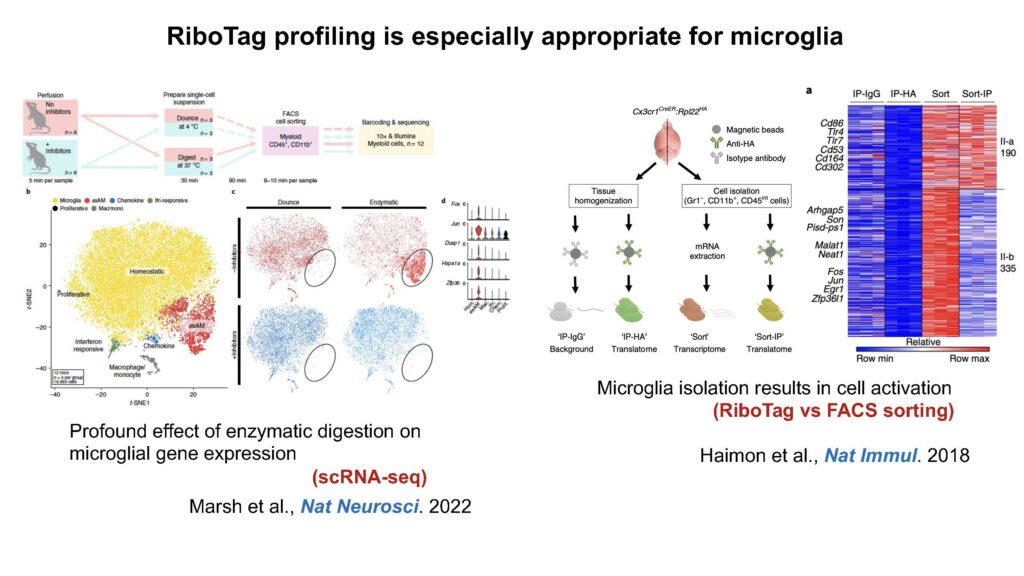
(4) RiboTag profiling
Microglia are highly sensitive and dynamic cells. I chose to use RiboTag profiling to address how loss of insulin signaling or concurrent loss of insulin signaling and AD pathogenesis could alter microglia translation.
Below I attached two slides I used to introduce what is RiboTag and why it is important for microglia profiling.
I also copied the written description on microglia RiboTag profiling from my submitted paper.
RiboTag isolation
4-month-old male MGRiboTag and MG-IRKORiboTag mice were deeply anesthetized and perfused using ice-cold PBS. The brains were separated and frozen on dry ice and stored at -80 ºC for storage. To extract microglial-specific ribosomal mRNA from bulk tissues, we performed RiboTag isolation as previously reported with modifications (Gao and Zhao, 2021; Mahadevan et al., 2020; Sanz et al., 2019). Briefly, brain hemispheres were homogenized in a pre-chilled Dounce filled in a freshly prepared iced homogenization buffer supplemented with 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 12 mM MgCl2, 1% NP40, 1 mM DTT, 1x protease inhibitor cocktail (B14002, Bimake), 200 U/mL RNasin (Promega), 100 µg/mL cycloheximide, and 1 mg/mL heparin. Five percent of each sample was separately prepared as Input samples, representing the whole transcriptome, while the remaining sample was proceeded for isolating ribosome-bound mRNA using RiboTag immunoprecipitation (IP) and redeemed as IP. mRNA from IP samples were subsequently isolated with a Direct-zol RNA Microprep Kit (R2062, Zymo).
(5) siRNA transfection
- siRNA-mediated knockdown of gene expression in cultured cells is designed for studying gene function and regulatory pathways.
- It usually includes two key steps: (1) siRNA design and preparation, (2) delivery using lipid-based transfection.
- For lipid-based transfection, cells are seeded and transfected with siRNA-lipid complexes, followed by incubation and assessment of knockdown efficiency.
- Post-transfection, knockdown efficiency is evaluated using RT-PCR for mRNA levels and Western blotting for protein levels.
- Functional assays (cell viability, migration, apoptosis, or pathway analysis) are usually performed to assess the impact of gene silencing.
- Critical considerations: (1) selecting appropriate controls, (2) optimizing siRNA concentration, (3) avoiding antibiotics during transfection, (4) confirming knockdown at both RNA and protein levels.
I also copied the written description on siRNA transinfection from my submitted paper.
siRNA Transinfection
SIM-A9 cells were seeded at a density of 1 × 105 cells cm−2 for 24 hrs prior to the transfection. The cells were transfected with 10 nM siRNA using Lipofectamine RNAiMAX (13778150, ThermoFisher) overnight in the plates according to manufacturer’s protocol. The next day in the morning, culture medium was changed to fresh maintenance medium for 72 hrs until experiments. SMARTpool ON-TARGETplus control siRNA (D-001810-01-20) and mouse Insr siRNA (L-043748-00-0005) were purchased from Horizon Discovery. The target sequences for each siRNA are provided in the Supplementary Table 1. Efficiency and specificity of siRNA transfection was verified using RT-qPCR.
I also attached the key reagent I have used.
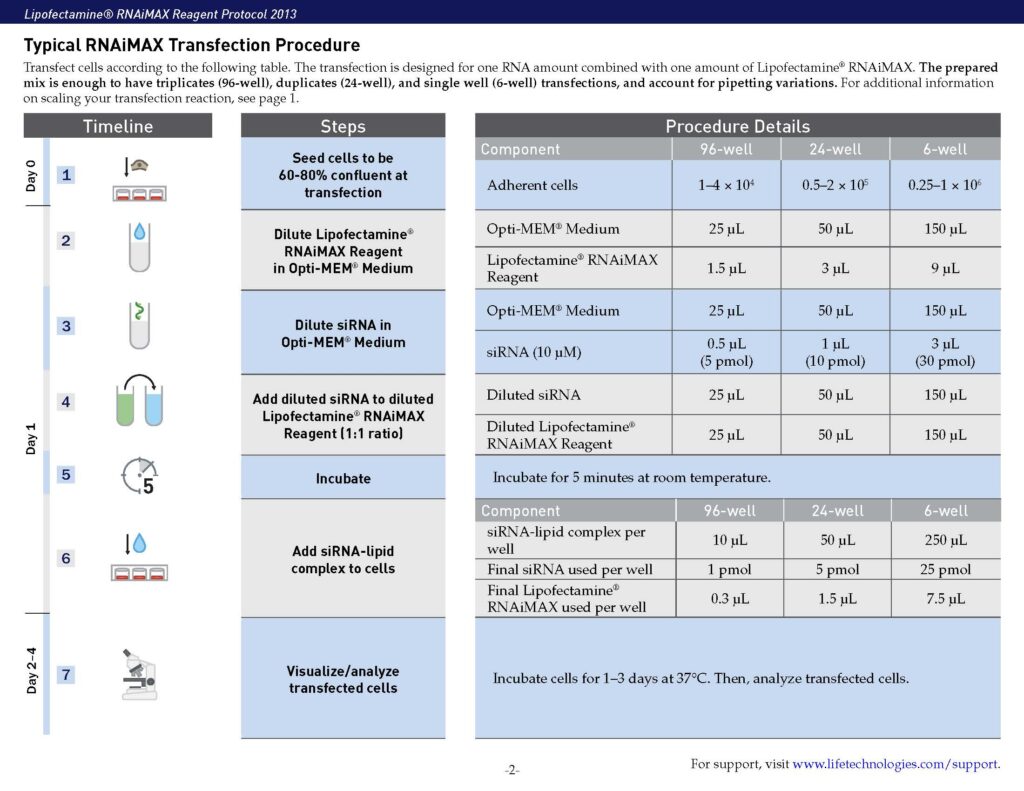
Further reading:
- https://pmc.ncbi.nlm.nih.gov/articles/PMC6743327/
- https://pmc.ncbi.nlm.nih.gov/articles/PMC8293861/
(6) Human iPSC-derived microglia
- Most of my work folloed Mathew Blurton-Jones’s publication (Neuron. 2017).
- Link: https://pubmed.ncbi.nlm.nih.gov/28426964/.
- Title: iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases.
- The key reagent I use is STEMCELL’s STEMdiff™ Hematopoietic Kit (Catalog # 05310).
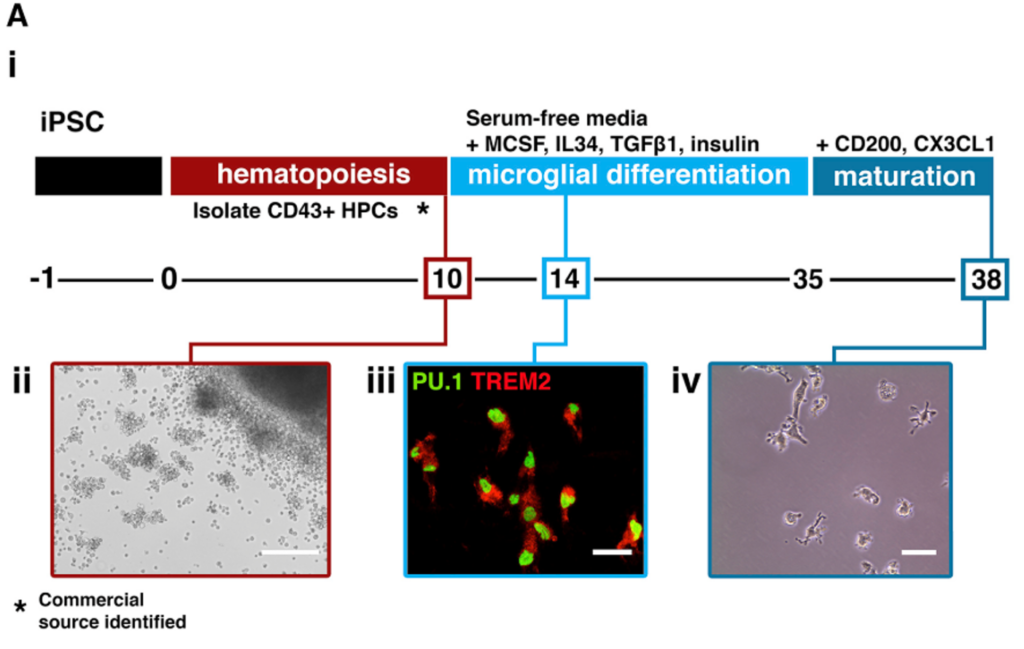
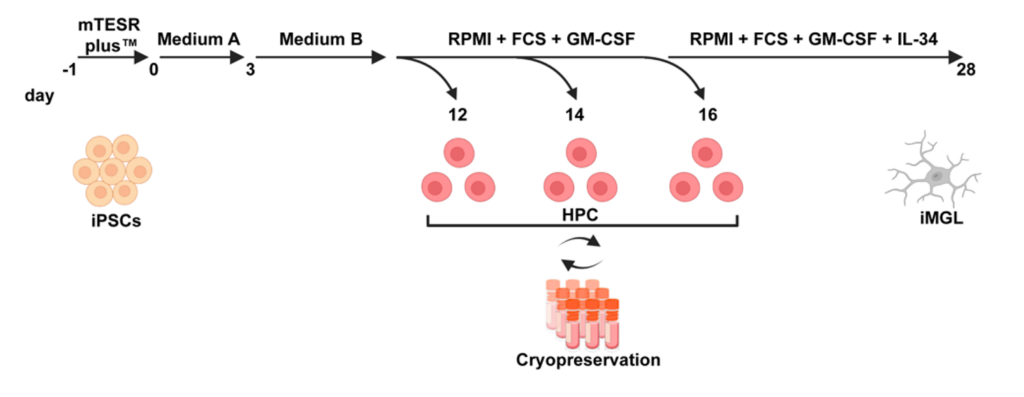
- There is a shorter protocol, which only needs 28 days to obtain matured microglia
- Link: https://www.mdpi.com/1422-0067/23/9/4526
- Title: Efficient and Easy Conversion of Human iPSCs into Functional Induced Microglia-like Cells.
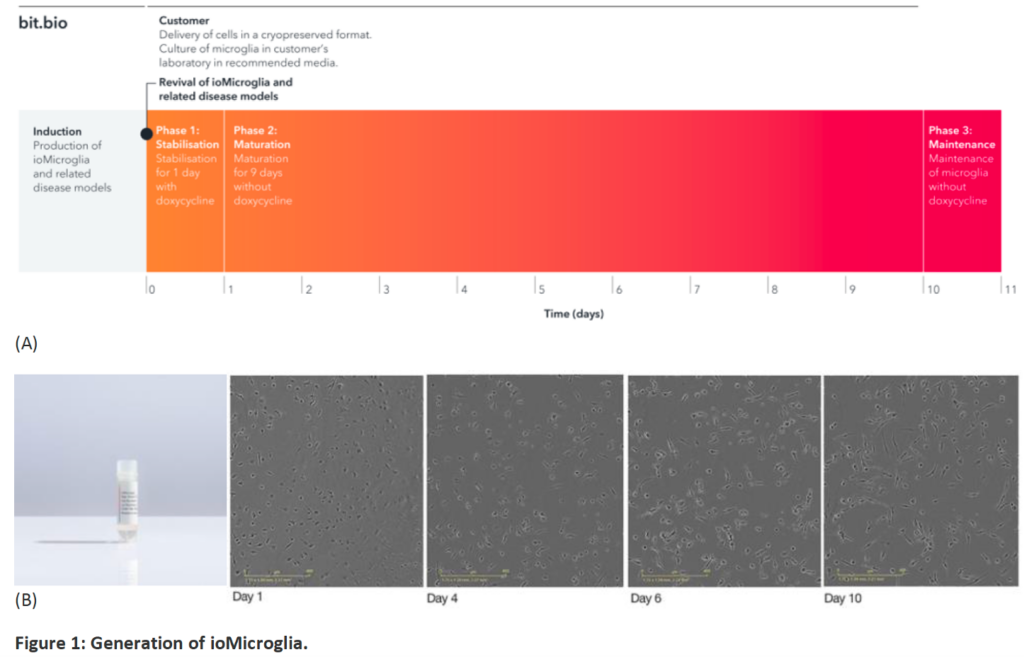
- to obtain disease microglia from human patients, I have purchased iPS-derived microglia from bit.bio, which has many disease backgrounds, AD, most importantly
- bit.bio mainly uses an approach called “opti-ox” (optimised inducible overexpression), which can force the cells to switch identity.
- Link: https://www.bit.bio/products/glial-cells/microglia-wild-type-io1021
- Cells: ioMicroglia Male Male human iPSC donor-derived microglia
- Once obtain the cells and thrawed from vials, it needs 10 days for the microglia maturation.
- On my hands, after 4-day maturation, these cells can already exhibit microglia-like morphology. See below a phase contrast picture from D4 (these iPS-derived cells are from AD humans with TREM2 R47H mutation)
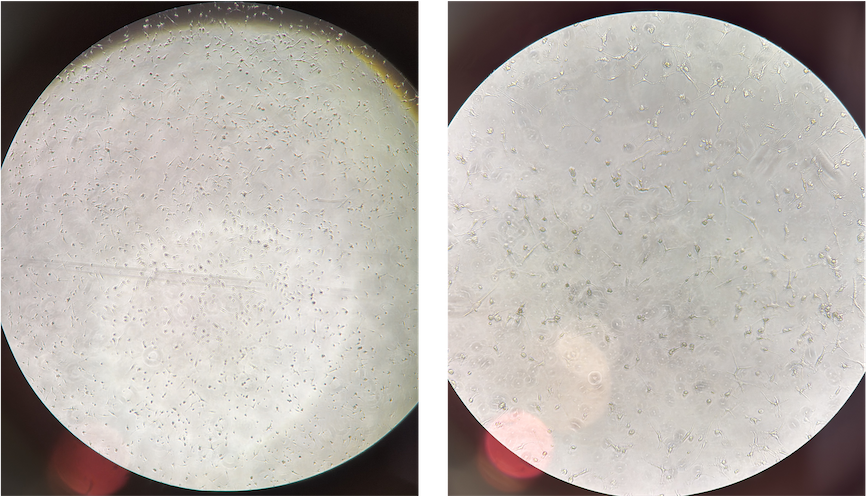
CRISPR-read iMicroglia
- These are human iPSC-derived microglial cells engineered to constitutively express Cas9 nuclease.
- I used it for a quick and easy generation of gene knockouts and CRISPR screens.
- Upon gRNA delivery from day 1 to 18 post-thaw, I can use them for KO experiment.
Example images
© 2026 All Rights Reserved.
Share:
More Posts
Global Collboration with Young PIs
Global Collaborationwith Young PIs My general statement: Do something, not be someone. Wenqiang Chen, Ph.D. As a young investigator in the field of neuroscience, I
Leadership
Leadership My general statement: Do something, not be someone. Wenqiang Chen, Ph.D. To me, leadership is one of the most essential qualities in any profession.
Mentorship – my Mentees
wenqiang.chen@joslin.harvard.edu wenqiang.chen.01@regionh.dk Teaching & my mentees My general statements: “Having been shaped by incredible mentors, I am committed to paying it forward. My goal is
Mentorship – my Mentors
wenqiang.chen@joslin.harvard.edu wenqiang.chen.01@regionh.dk Mentorship My general statements: – Research lets me explore science and teach it. – Great scientists should also be great mentors. – Great
